
Zhen Qian, Junzhou Huang, Xiaolei Huang, Zhiguo Li, Dimitris Metaxas, Leon Axel
IntroductionHeart failure is a leading cause of morbidity and mortality. Consequently, the study of normal and
pathological heart behavior is the topic of rigorous research. In particular the study of the shape and
motion of the heart is important because these factors are thought to strongly correlate with many heart
diseases. For instance, alteration of heart wall motion is a sensitive indicator of heart disease; moreover, abnormalities in heart wall motion
are taken very seriously by physicians, because they can be life threatening.
There are two important research questions in assessing heart wall motion: 1) how do we investigate the motion and mechanics of the beating
heart without significant perturbation of its function, and 2) how do we quantitatively extract heart wall motion parameters that capture the
differences between normal and diseased states, and in a way that is useful to physicians. The first question is partly answered by the
development of non-invasive imaging technologies such as Magnetic Resonance Imaging (MRI), ultrasound, and Computed Tomography (CT).
Physicians are increasingly relying on these images for diagnosis, prognosis and treatment planning.
In order to utilize the generated images to better understand the complex regional cardiac mechanical changes under pathological conditions,
it is important to answer the second question of quantitative and proper characterization of regional heart wall motion. Researchers have used
tagged MRI images for precise, quantitative study of regional cardiac mechanics, however, this analysis is a time-consuming and laborious
manual process, which is preventing tagged MR from being used for routine clinical evaluation.
To solve this problem, in our research, first we present a novel framework for learning a joint shape and appearance model from a large set
of un-labelled training examples in arbitrary positions and orientations. The shape and intensity spaces are unified by implicitly representing
shapes as “images” in the space of distance transforms. A stochastic chord-based matching algorithm is developed to align photo-realistic
training examples under a common reference frame. Then dense local deformation fields, represented using the cubic B-spline based Free Form
Deformations (FFD), are recovered to register the training examples in both shape and intensity spaces. Principal Component Analysis (PCA)
is applied on the FFD control lattices to capture the variations in shape as well as on registered object interior textures.
It has been noted by several researchers that the rate-limiting step which prevents tagged MR from clinical use is the robust extraction and
tracking of the contours and tags. There have been a vast research efforts on the automated contour segmentation, however, it still remains a
difficult task due to the common presence of cluttered objects, complex object textures, image noise, intensity inhomogeneity, and especially
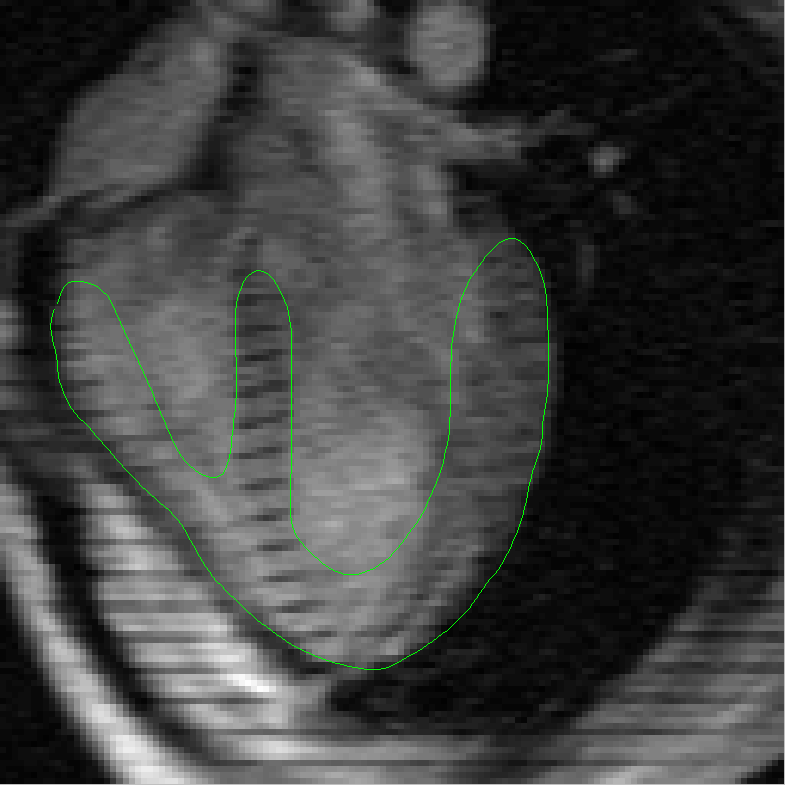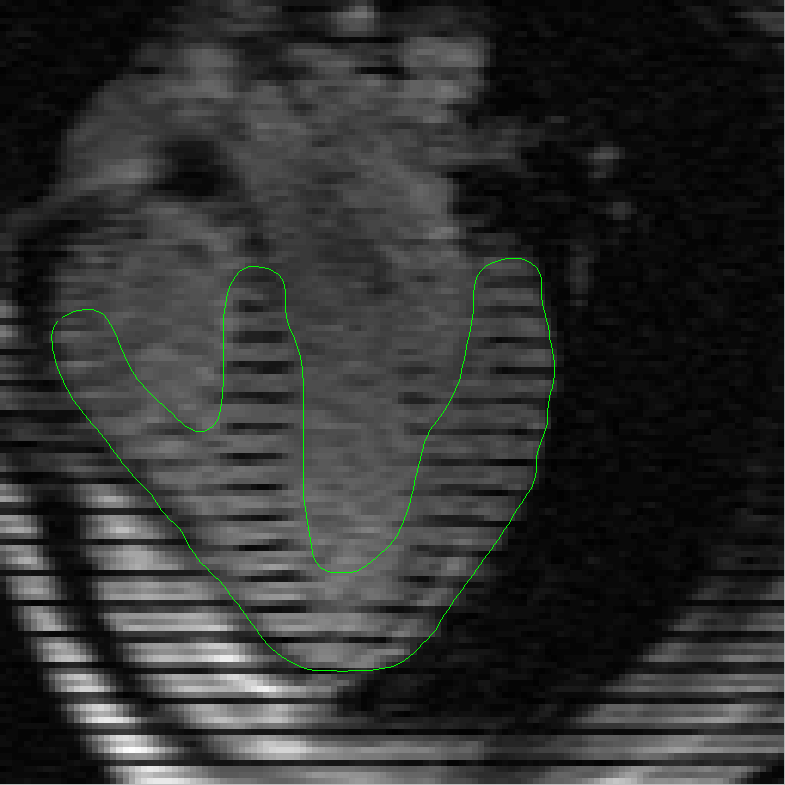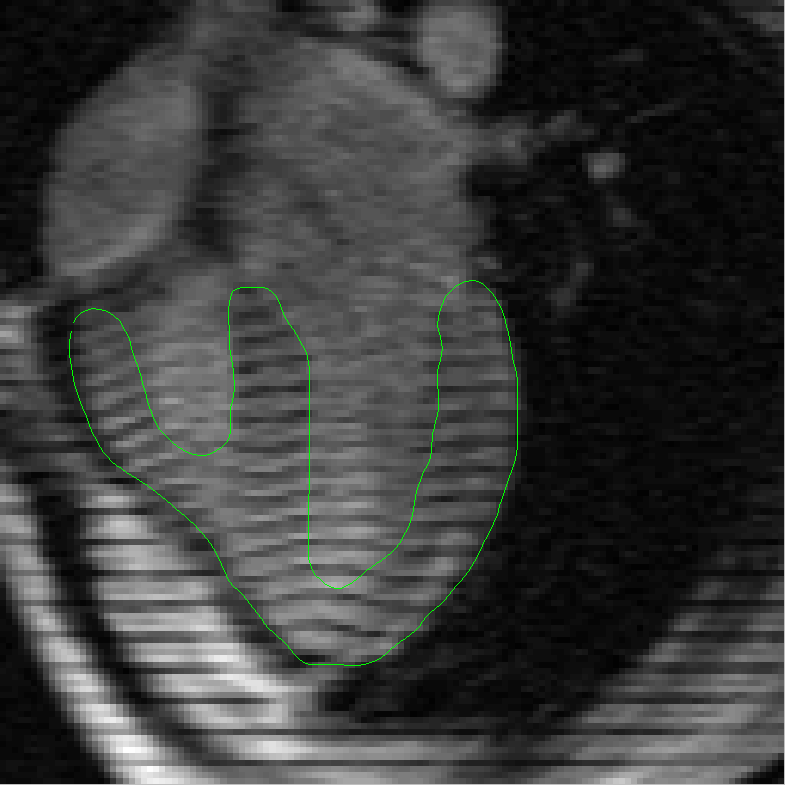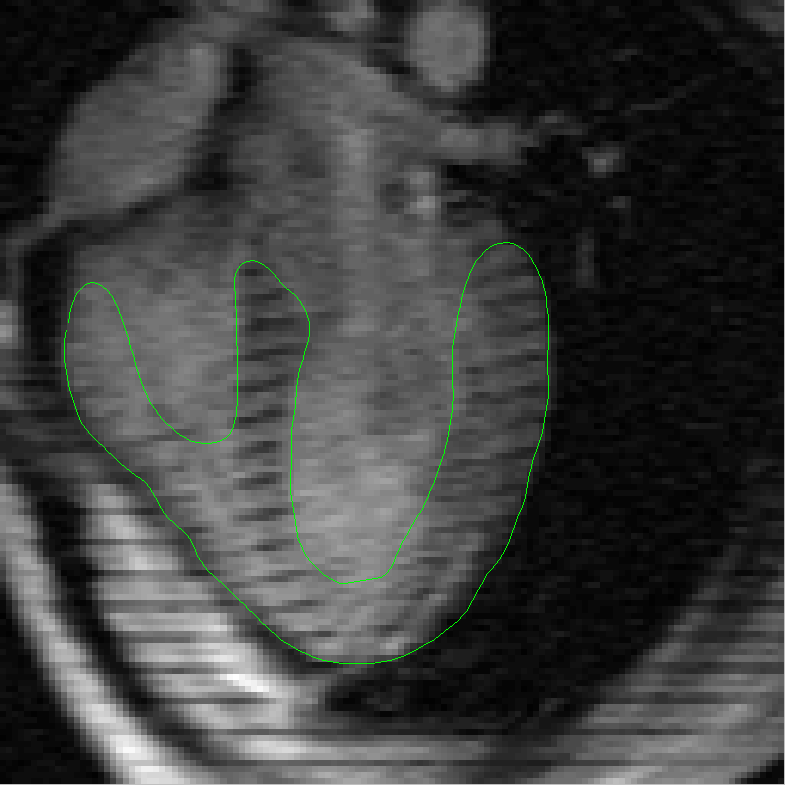
the complexities added by the tagging lines. In order to address this problem we then present a robust method for segmenting and tracking
cardiac contours and tags in 4D cardiacMRI tagged images via spatio-temporal propagation. Our method is based on two main techniques: the
Metamorphs Segmentation for robust boundary estimation, and the tunable Gabor filter bank for tagging lines enhancement, removal and
myocardium tracking. Our prototype system based on the integration of these two techniques achieved efficient, robust segmentation and
tracking with minimal human interaction.
The challenging problem of heart localization and segmentation in 4D Spatio-temporal cardiac images is also a problem that we want to address.
We furtherly present a dynamic texture based motion segmentation approach. Our approach introduces time-dependent dynamic constraints into
model-based segmentation, and it has the advantage of producing segmentation results that are both spatially and temporally consistent.
Compared with previous methods that segment cardiac contours, our method offers the following advantages: 1) with distinct dynamic signatures,
the heart can be quickly localized in 4D cardiac images; 2)heart dynamics are learned online and adaptively by analyzing the dynamic texture
from the video sequence of a cardiac cycle and then incorporated in the segmentation process; and 3) the proposed dynamic features can be
easily integrated with model-based segmentation methods.
In order to get better cardiac boundary segmentation and tag tracking, we introduce a tag separation method which is based on two observations
in the cardiac tagged MR images: 1) the tag patterns have a regular texture; 2) the cardiac images without tag patterns are piecewise smooth
with sparse gradients. These observations motivate us to use two dictionaries, one based on the Discrete Cosine Transform for representing
tag patterns and the other based on the Wavelet Transform for representing the underlying cardiac image without tag patterns. The two
dictionaries are built such that they can lead to sparse representations of the tag patterns and of the piece-wise smooth regions without
tag patterns. With the two dictionaries, a new tag separation approach is proposed to simultaneously optimize w.r.t. the two sparse
representations, where optimization is directed by the Total Variation regularization scheme. While previous methods have focused on tag
removal, our approach to acquiring both optimally-decomposed tag-only image and the cardiac image without tags simultaneously can be used
for better tag tracking and cardiac boundary segmentation.